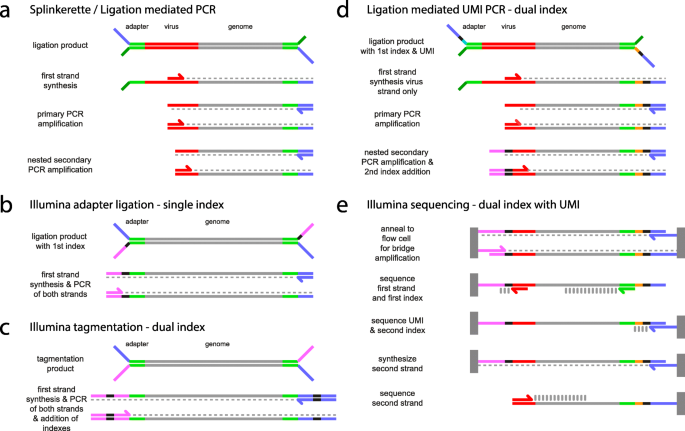
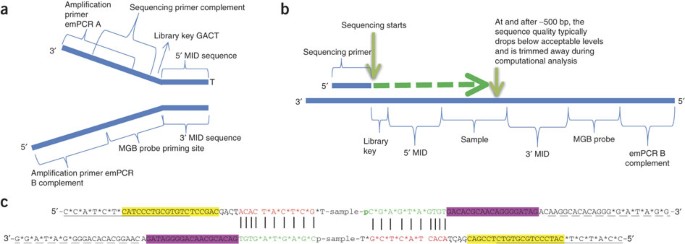
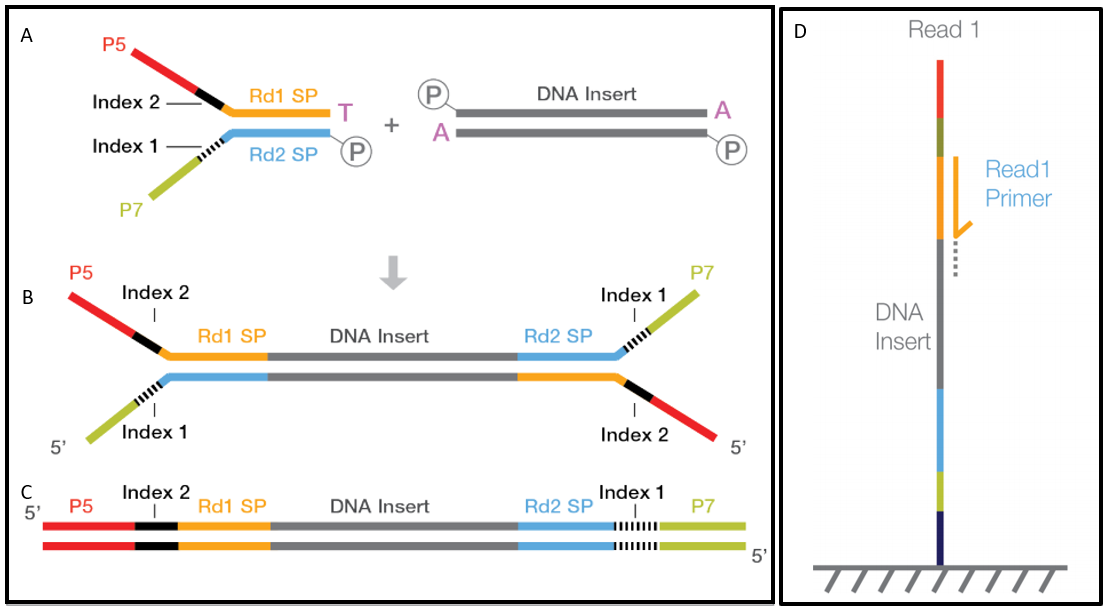
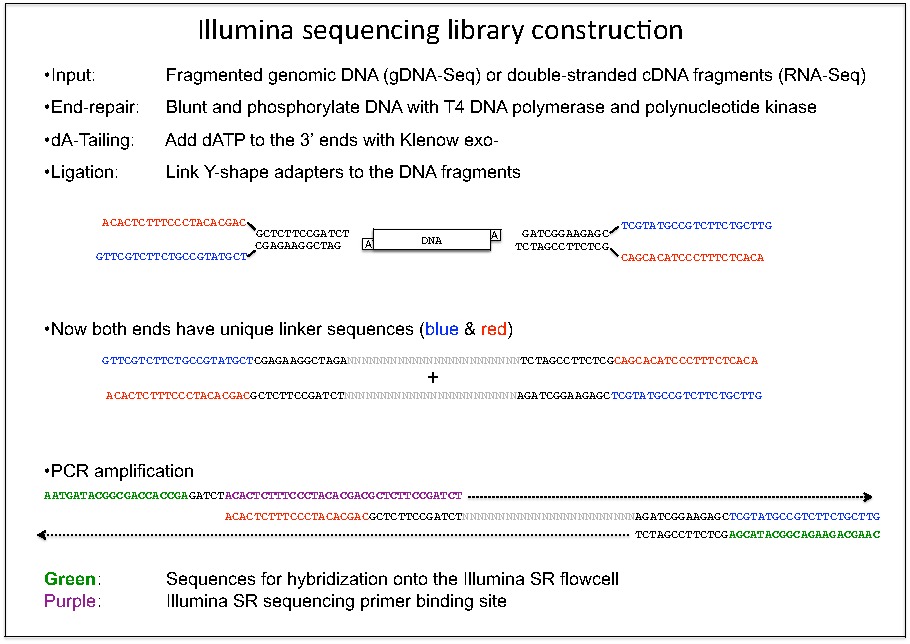
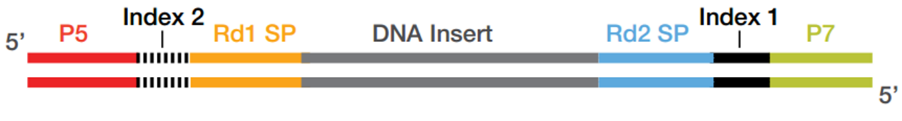
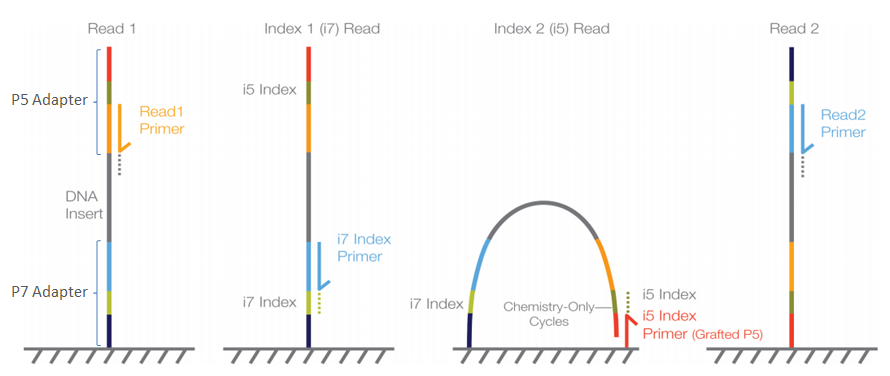
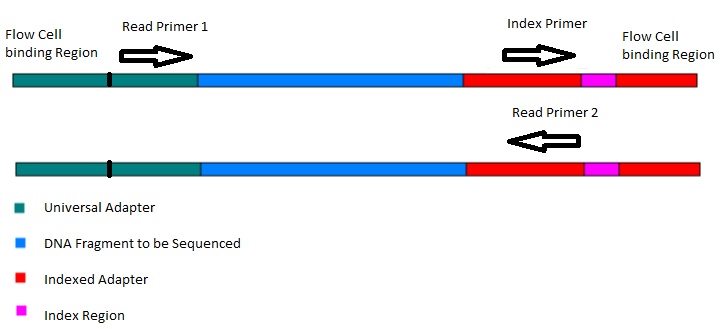
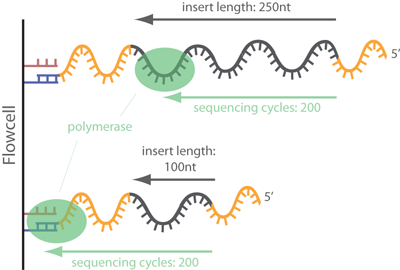
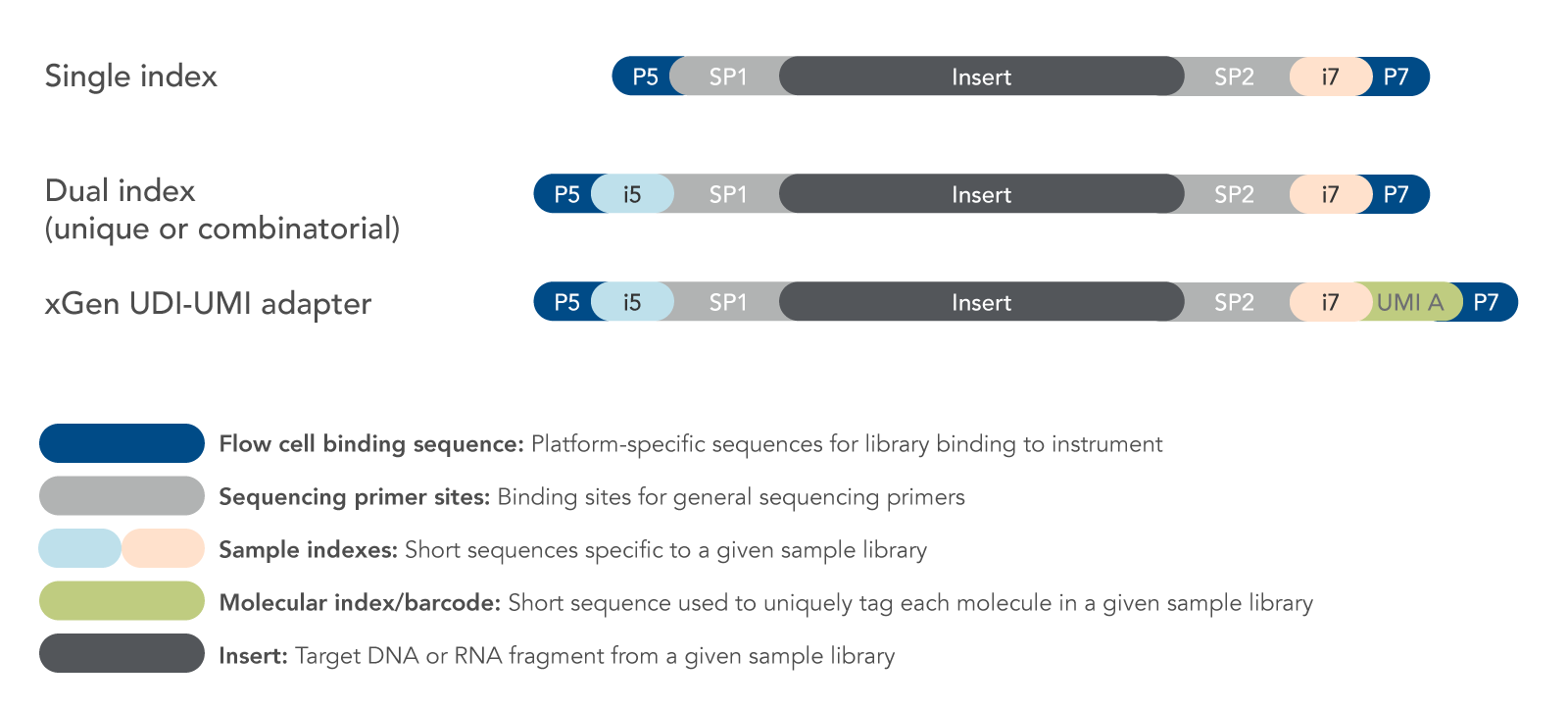
High-Throughput, Amplicon-Based Sequencing of the CREBBP Gene as a Tool to Develop a Universal Platform-Independent Assay | PLOS ONE

Adaptors and adaptor chimeras are a common sources of sequence artifacts | Download Scientific Diagram
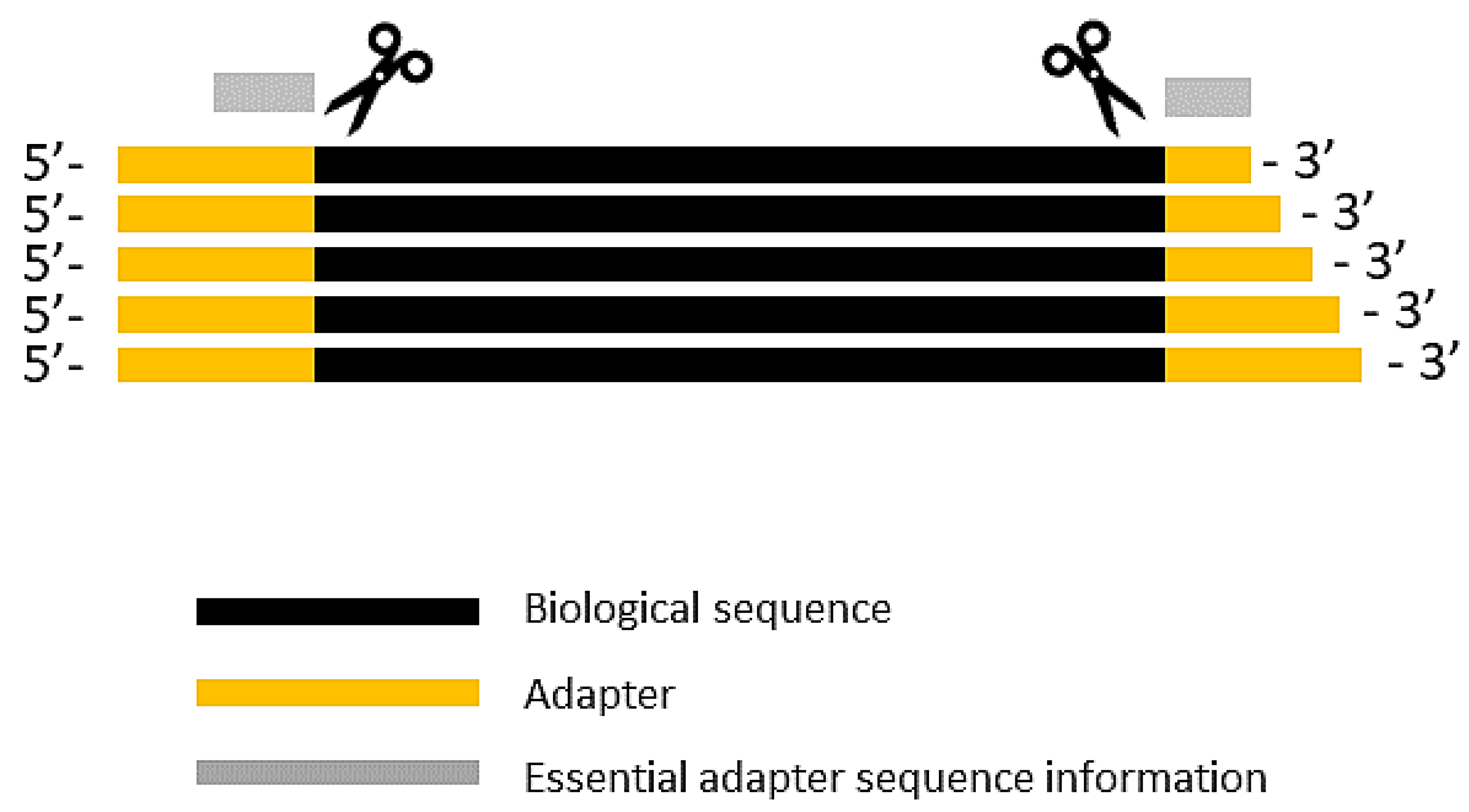
Biomolecules | Free Full-Text | High-Throughput Identification of Adapters in Single-Read Sequencing Data | HTML
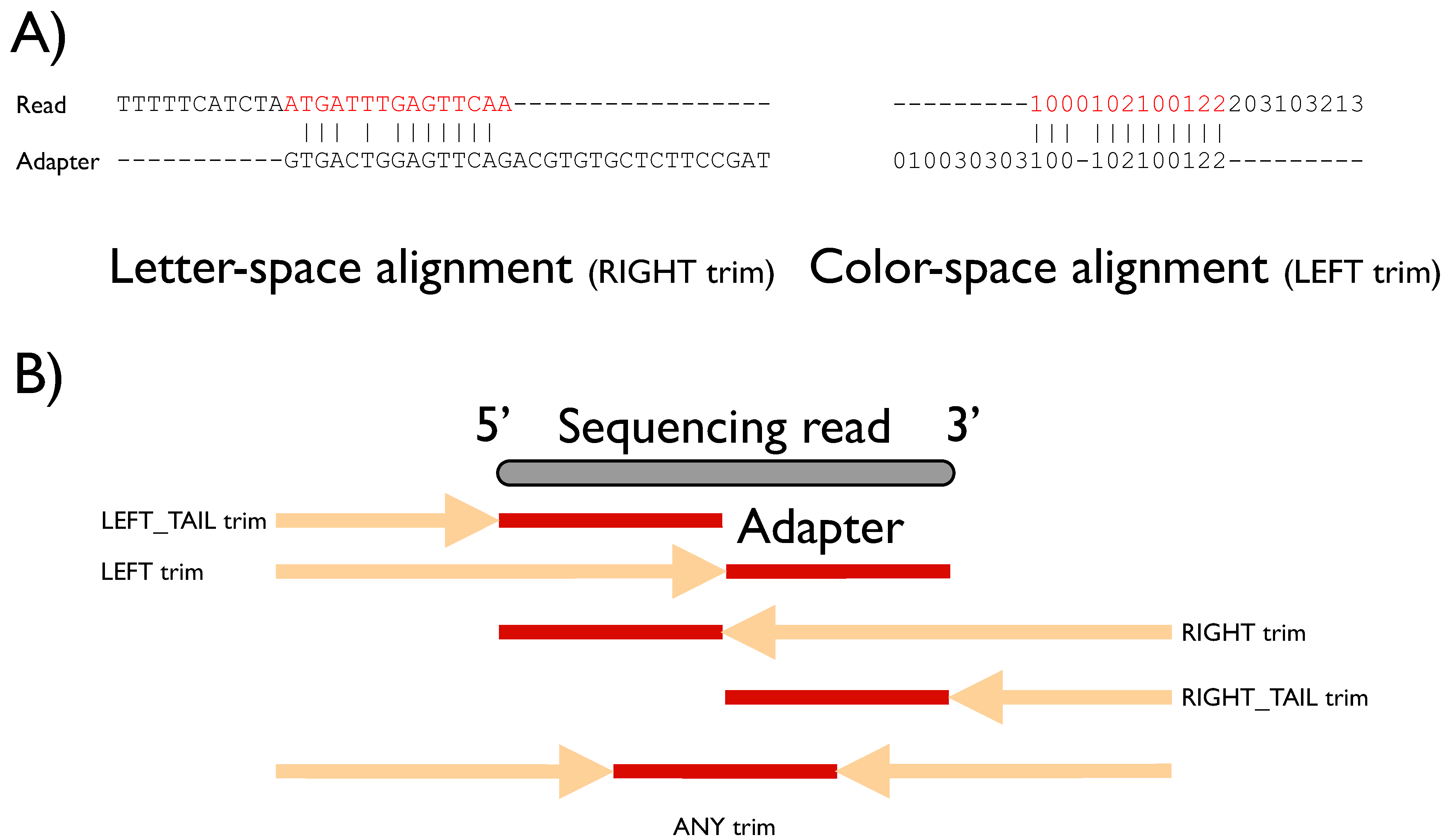
Biology | Free Full-Text | FLEXBAR—Flexible Barcode and Adapter Processing for Next-Generation Sequencing Platforms | HTML
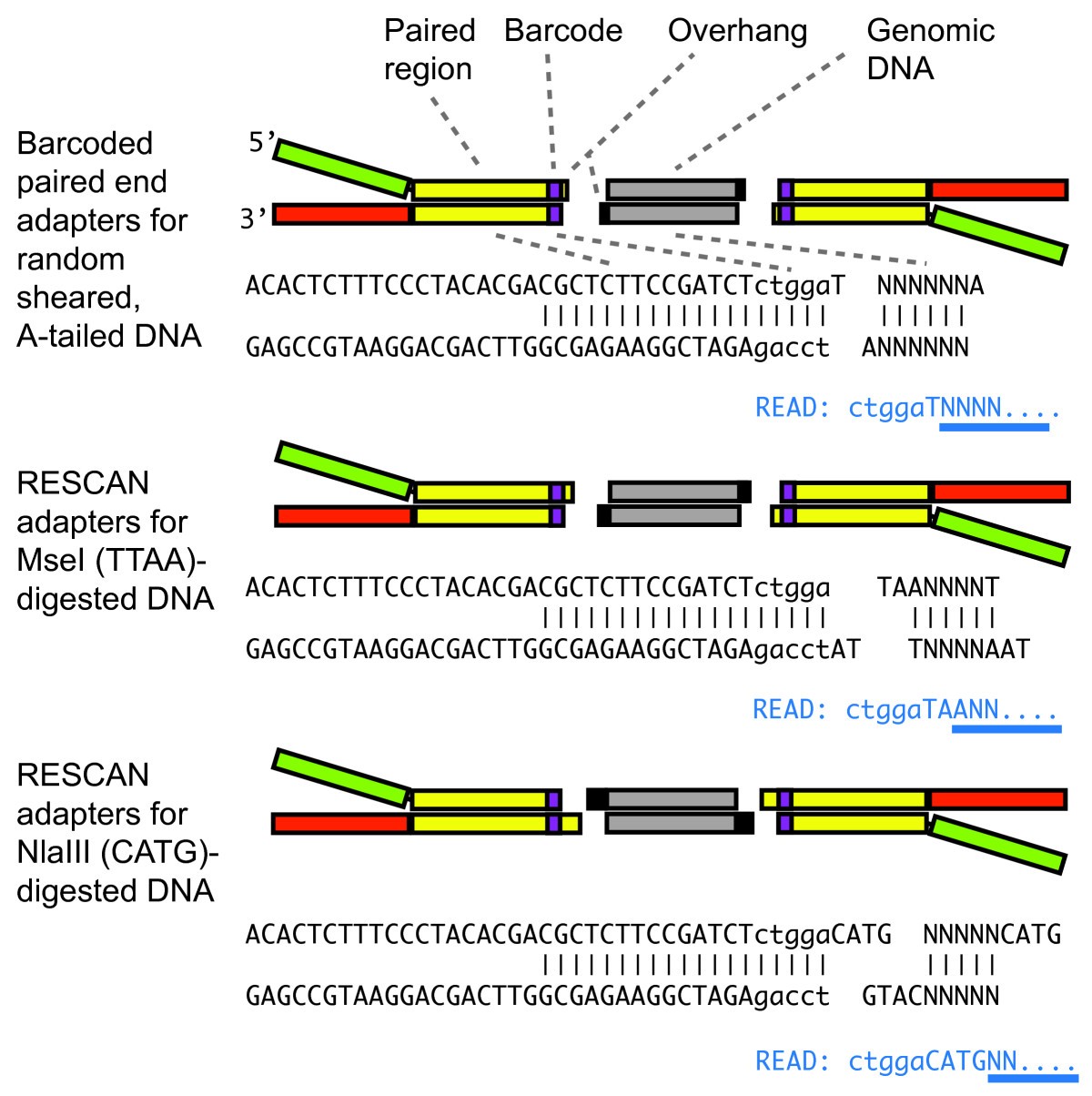
Reference genome-independent assessment of mutation density using restriction enzyme-phased sequencing | BMC Genomics | Full Text

Illumina Genome Analyzer sequencing. Adapter-modified, single-stranded... | Download Scientific Diagram
4-A scheme of targeted fragment being sequenced Sequencing adapters... | Download High-Quality Scientific Diagram